PyCaret Tutorial
PyCaret Tutorial
This tutorial will not cover PyCaret as a whole, but it will rather focus on the entire pipeline for a single classification problem. We will use the wine recognition dataset:
Lichman, M. (2013). UCI Machine Learning Repository. Irvine, CA: University of California, School of Information and Computer Science.
OBS: This post was automatically generated from my Jupyter Notebook thanks to Adam Blomberg’s post.
What is PyCaret?
PyCaret is a library that comes to automate the Machine Learning (ML) process requiring very few lines of code. It works with multiple other useful ML libraries, such as scikit-learn.
Check out their website for more information: PyCaret Homepage.
Installing PyCaret
Virtual environment
First, we create a virtual environment to ensure we do not have unnecessary packages as well as to keep things isolated and light.
Using venv:
$ python -m venv /path/to/new/virtual/environment
And to activate it:
$ source /path/to/new/virtual/environment/bin activate
Using conda:
$ conda create --name venv pip
And to activate it:
$ conda source venv activate
Actual PyCaret installation
We use pip to install it:
$ pip install pycaret
Or on the notebook:
! pip install pycaret
Dataset
We will import the dataset from Scikit-learn:
import sklearn
from sklearn.datasets import load_wine
print(f"Scikit-learn version: {sklearn.__version__}")
import pandas as pd
import numpy as np
Scikit-learn version: 0.23.2
data = load_wine()
df = pd.DataFrame(data=np.c_[data['data'], data['target']],
columns=data['feature_names'] + ['target'])
df.shape
(178, 14)
Let’s do some very brief EDA on the dataset:
display(df.head())
df.describe()
| alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280/od315_of_diluted_wines | proline | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14.23 | 1.71 | 2.43 | 15.6 | 127.0 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065.0 | 0.0 |
| 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100.0 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050.0 | 0.0 |
| 2 | 13.16 | 2.36 | 2.67 | 18.6 | 101.0 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185.0 | 0.0 |
| 3 | 14.37 | 1.95 | 2.50 | 16.8 | 113.0 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480.0 | 0.0 |
| 4 | 13.24 | 2.59 | 2.87 | 21.0 | 118.0 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735.0 | 0.0 |
| alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280/od315_of_diluted_wines | proline | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 | 178.000000 |
| mean | 13.000618 | 2.336348 | 2.366517 | 19.494944 | 99.741573 | 2.295112 | 2.029270 | 0.361854 | 1.590899 | 5.058090 | 0.957449 | 2.611685 | 746.893258 | 0.938202 |
| std | 0.811827 | 1.117146 | 0.274344 | 3.339564 | 14.282484 | 0.625851 | 0.998859 | 0.124453 | 0.572359 | 2.318286 | 0.228572 | 0.709990 | 314.907474 | 0.775035 |
| min | 11.030000 | 0.740000 | 1.360000 | 10.600000 | 70.000000 | 0.980000 | 0.340000 | 0.130000 | 0.410000 | 1.280000 | 0.480000 | 1.270000 | 278.000000 | 0.000000 |
| 25% | 12.362500 | 1.602500 | 2.210000 | 17.200000 | 88.000000 | 1.742500 | 1.205000 | 0.270000 | 1.250000 | 3.220000 | 0.782500 | 1.937500 | 500.500000 | 0.000000 |
| 50% | 13.050000 | 1.865000 | 2.360000 | 19.500000 | 98.000000 | 2.355000 | 2.135000 | 0.340000 | 1.555000 | 4.690000 | 0.965000 | 2.780000 | 673.500000 | 1.000000 |
| 75% | 13.677500 | 3.082500 | 2.557500 | 21.500000 | 107.000000 | 2.800000 | 2.875000 | 0.437500 | 1.950000 | 6.200000 | 1.120000 | 3.170000 | 985.000000 | 2.000000 |
| max | 14.830000 | 5.800000 | 3.230000 | 30.000000 | 162.000000 | 3.880000 | 5.080000 | 0.660000 | 3.580000 | 13.000000 | 1.710000 | 4.000000 | 1680.000000 | 2.000000 |
All data is numeric and without missing values.
# Checking the classes
df['target'].value_counts()
1.0 71
0.0 59
2.0 48
Name: target, dtype: int64
The classes are not very imbalanced.
Splitting the dataset into train and test
from sklearn.model_selection import train_test_split
df_train, df_test = train_test_split(df,
test_size=0.20,
random_state=0,
stratify=df['target'])
Initializing PyCaret
We can, now, import PyCaret and start playing around with it!
import pycaret
from pycaret.classification import *
print(f"PyCaret version: {pycaret.__version__}")
PyCaret version: 2.3.1
PyCaret syntax asks us to setup the data as the first step, providing the input data with the features and the target variable. This setup function is how PyCaret initializes the pipeline for future preprocessing, modeling, and deployment. It requires two parameters: data, a pandas dataframe, and target, the name of the target column. There are other parameters, but they are all optional and we will not cover them at this point.
The setup infers on its own the features’ data types, but it is worth double checking since it may do it incorrectly sometimes. A table showing the features and their data types is displayed after running the setup. It asks us to confirm if correct and press enter. If something is incorrect, then we can correct it by typing quit. Otherwise, we can proceed. Notice that getting the data types right is extremely important, because PyCaret automatically preprocesses the data based on each feature’s type. Preprocessing is a fundamental and critical part of doing ML properly.
# setup the dataset
grid = setup(data=df_train, target='target')
| Description | Value | |
|---|---|---|
| 0 | session_id | 6420 |
| 1 | Target | target |
| 2 | Target Type | Multiclass |
| 3 | Label Encoded | None |
| 4 | Original Data | (142, 14) |
| 5 | Missing Values | False |
| 6 | Numeric Features | 13 |
| 7 | Categorical Features | 0 |
| 8 | Ordinal Features | False |
| 9 | High Cardinality Features | False |
| 10 | High Cardinality Method | None |
| 11 | Transformed Train Set | (99, 13) |
| 12 | Transformed Test Set | (43, 13) |
| 13 | Shuffle Train-Test | True |
| 14 | Stratify Train-Test | False |
| 15 | Fold Generator | StratifiedKFold |
| 16 | Fold Number | 10 |
| 17 | CPU Jobs | -1 |
| 18 | Use GPU | False |
| 19 | Log Experiment | False |
| 20 | Experiment Name | clf-default-name |
| 21 | USI | 6d1b |
| 22 | Imputation Type | simple |
| 23 | Iterative Imputation Iteration | None |
| 24 | Numeric Imputer | mean |
| 25 | Iterative Imputation Numeric Model | None |
| 26 | Categorical Imputer | constant |
| 27 | Iterative Imputation Categorical Model | None |
| 28 | Unknown Categoricals Handling | least_frequent |
| 29 | Normalize | False |
| 30 | Normalize Method | None |
| 31 | Transformation | False |
| 32 | Transformation Method | None |
| 33 | PCA | False |
| 34 | PCA Method | None |
| 35 | PCA Components | None |
| 36 | Ignore Low Variance | False |
| 37 | Combine Rare Levels | False |
| 38 | Rare Level Threshold | None |
| 39 | Numeric Binning | False |
| 40 | Remove Outliers | False |
| 41 | Outliers Threshold | None |
| 42 | Remove Multicollinearity | False |
| 43 | Multicollinearity Threshold | None |
| 44 | Clustering | False |
| 45 | Clustering Iteration | None |
| 46 | Polynomial Features | False |
| 47 | Polynomial Degree | None |
| 48 | Trignometry Features | False |
| 49 | Polynomial Threshold | None |
| 50 | Group Features | False |
| 51 | Feature Selection | False |
| 52 | Feature Selection Method | classic |
| 53 | Features Selection Threshold | None |
| 54 | Feature Interaction | False |
| 55 | Feature Ratio | False |
| 56 | Interaction Threshold | None |
| 57 | Fix Imbalance | False |
| 58 | Fix Imbalance Method | SMOTE |
Among the optional parameters we skipped, there were options for preprocessing such as outline removal, feature selection, feature encoding, dimensionality reduction, how to split between train and test set, and many more! Check out the documentation for more details.
Comparing different models
After setting up, the time to compare and evaluate different models has arrived! And this is done so easily that pretty much anyone with a minimal knowledge on metrics could pick up the best model. This is one of the cool PyCaret’s features that allows us to save a lot of time.
PyCaret uses 10-fold cross-validation as its default, sort results by classification accuracy, returns the best model, and displays the results of all tested classifiers with different metrics.
best = compare_models()
| Model | Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | TT (Sec) | |
|---|---|---|---|---|---|---|---|---|---|
| rf | Random Forest Classifier | 0.9700 | 0.9975 | 0.9750 | 0.9770 | 0.9698 | 0.9545 | 0.9585 | 0.0790 |
| nb | Naive Bayes | 0.9600 | 0.9950 | 0.9639 | 0.9678 | 0.9598 | 0.9394 | 0.9431 | 0.0060 |
| et | Extra Trees Classifier | 0.9600 | 0.9967 | 0.9700 | 0.9703 | 0.9604 | 0.9384 | 0.9438 | 0.0670 |
| ridge | Ridge Classifier | 0.9589 | 0.0000 | 0.9600 | 0.9686 | 0.9578 | 0.9365 | 0.9423 | 0.0050 |
| lightgbm | Light Gradient Boosting Machine | 0.9500 | 1.0000 | 0.9589 | 0.9623 | 0.9486 | 0.9242 | 0.9315 | 0.1100 |
| lda | Linear Discriminant Analysis | 0.9489 | 0.9936 | 0.9533 | 0.9619 | 0.9483 | 0.9211 | 0.9283 | 0.0060 |
| qda | Quadratic Discriminant Analysis | 0.9389 | 0.9950 | 0.9167 | 0.9534 | 0.9319 | 0.9023 | 0.9135 | 0.0070 |
| dt | Decision Tree Classifier | 0.9200 | 0.9369 | 0.9167 | 0.9330 | 0.9178 | 0.8773 | 0.8855 | 0.0060 |
| lr | Logistic Regression | 0.9189 | 0.9888 | 0.9194 | 0.9314 | 0.9175 | 0.8762 | 0.8830 | 0.4420 |
| gbc | Gradient Boosting Classifier | 0.8900 | 0.9876 | 0.8922 | 0.9127 | 0.8883 | 0.8290 | 0.8416 | 0.0770 |
| ada | Ada Boost Classifier | 0.8489 | 0.9378 | 0.8378 | 0.8721 | 0.8440 | 0.7652 | 0.7782 | 0.0320 |
| knn | K Neighbors Classifier | 0.7178 | 0.8830 | 0.6922 | 0.7521 | 0.7012 | 0.5651 | 0.5915 | 0.0090 |
| svm | SVM - Linear Kernel | 0.5978 | 0.0000 | 0.5678 | 0.5420 | 0.5152 | 0.3702 | 0.4458 | 0.0070 |
Showing the best classifier:
print(best)
RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=-1, oob_score=False, random_state=6420, verbose=0,
warm_start=False)
Tuning a model
The models used when comparing use their default hyperparameters. We can tune and choose the best hyperparameters for a single model using PyCaret. This is done by running a Random Grid Search on a specific search space. We can, as well, define a custom search grid, but we will not do it at this point. Also, there are many hyperparameters that allow us to choose early stopping, number of iterations, which metric to optimize for, and so on. Tuning returns a similar table as before, but each row now shows the result for each validation fold.
We will tune the K Neighbors Classifier, since it performed poorly with its default parameters. To do so, we first create a model with PyCaret:
from sklearn.neighbors import KNeighborsClassifier
tuned_knn = tune_model(KNeighborsClassifier(), n_iter=100)
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 1 | 0.9000 | 0.9300 | 0.9333 | 0.9250 | 0.9016 | 0.8438 | 0.8573 |
| 2 | 0.7000 | 0.7929 | 0.6944 | 0.7400 | 0.7067 | 0.5385 | 0.5471 |
| 3 | 0.8000 | 0.9083 | 0.8056 | 0.8250 | 0.7971 | 0.6970 | 0.7078 |
| 4 | 0.9000 | 0.9012 | 0.8889 | 0.9200 | 0.8956 | 0.8462 | 0.8598 |
| 5 | 0.8000 | 0.8298 | 0.7778 | 0.8450 | 0.7627 | 0.6923 | 0.7273 |
| 6 | 0.7000 | 0.8857 | 0.6944 | 0.7400 | 0.7067 | 0.5385 | 0.5471 |
| 7 | 0.9000 | 1.0000 | 0.9167 | 0.9333 | 0.9029 | 0.8485 | 0.8616 |
| 8 | 0.8000 | 0.8458 | 0.8333 | 0.9000 | 0.8000 | 0.7059 | 0.7500 |
| 9 | 0.7778 | 0.9000 | 0.6667 | 0.6296 | 0.6889 | 0.6250 | 0.6934 |
| Mean | 0.8278 | 0.8994 | 0.8211 | 0.8458 | 0.8162 | 0.7335 | 0.7551 |
| SD | 0.0910 | 0.0637 | 0.1079 | 0.1076 | 0.0993 | 0.1417 | 0.1364 |
print(tuned_knn)
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='manhattan',
metric_params=None, n_jobs=None, n_neighbors=3, p=2,
weights='distance')
Plotting a model
If just seeing a table is not enough, you can plot the results of a model in different ways. Here are a few examples:
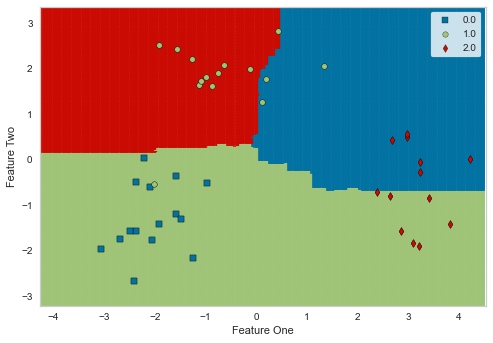
plot_model(best, plot='boundary')
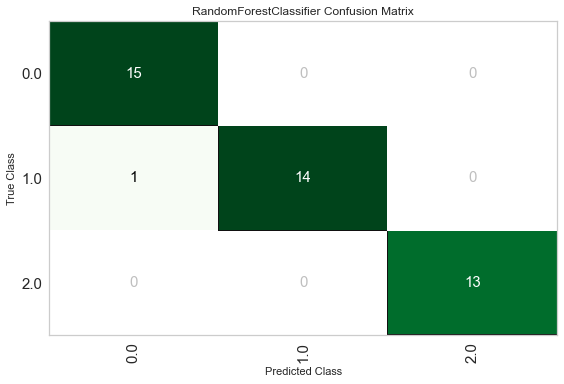
plot_model(best, plot='confusion_matrix')
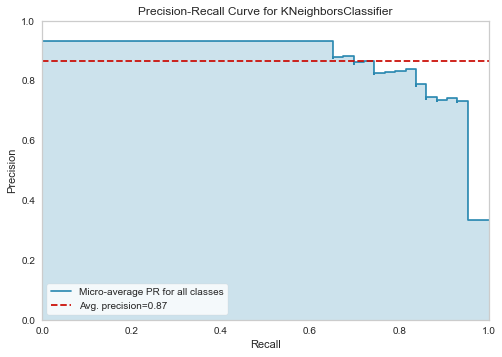
plot_model(tuned_knn, plot='pr')
plot_model(tuned_knn, plot='class_report')

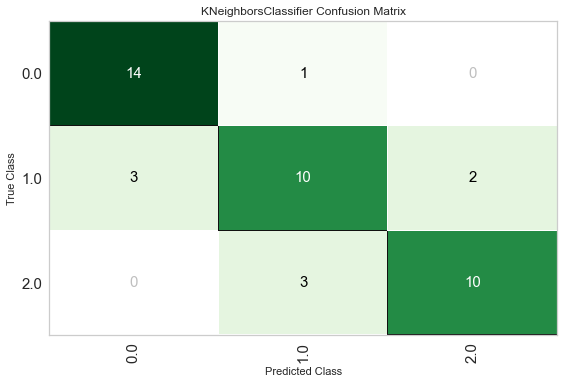
plot_model(tuned_knn, plot='confusion_matrix')
plot_model(tuned_knn, plot='auc')
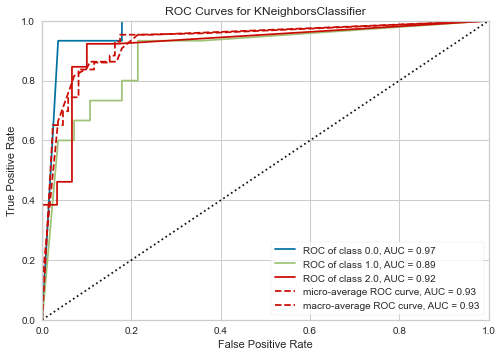
Explainable AI
Most businesses do not like to have a black-box model telling them what to do. It is extremely important to understand what the model does so that the business can take action to improve their results.
For this, we need to install the shap library:
$ pip install shap
Or on the notebook:
! pip install shap
This library is based on the concept of Shapley values created in the Game Theory context to compute feature importance. For now, it is only available for tree-based models.
from sklearn.ensemble import RandomForestClassifier
rf = create_model(RandomForestClassifier())
| Accuracy | AUC | Recall | Prec. | F1 | Kappa | MCC | |
|---|---|---|---|---|---|---|---|
| 0 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 1 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 2 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 3 | 0.9000 | 1.0000 | 0.9167 | 0.9250 | 0.9000 | 0.8507 | 0.8636 |
| 4 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 5 | 0.9000 | 1.0000 | 0.9167 | 0.9250 | 0.9000 | 0.8507 | 0.8636 |
| 6 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 7 | 0.9000 | 0.9667 | 0.9167 | 0.9200 | 0.8984 | 0.8438 | 0.8573 |
| 8 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 9 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Mean | 0.9700 | 0.9967 | 0.9750 | 0.9770 | 0.9698 | 0.9545 | 0.9585 |
| SD | 0.0458 | 0.0100 | 0.0382 | 0.0352 | 0.0461 | 0.0695 | 0.0635 |
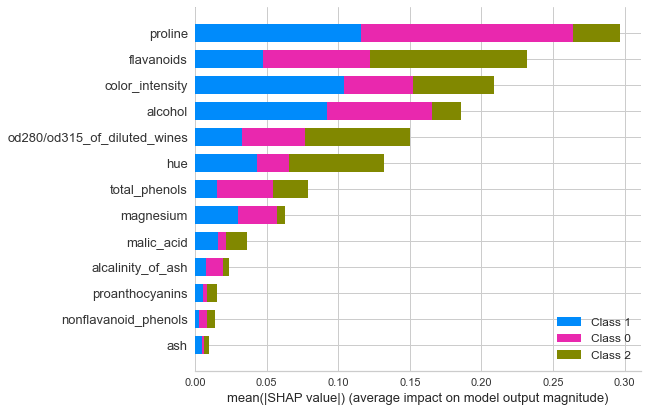
Summary plot
interpret_model(rf, plot='summary')
Correlation plot
interpret_model(rf, plot='correlation')

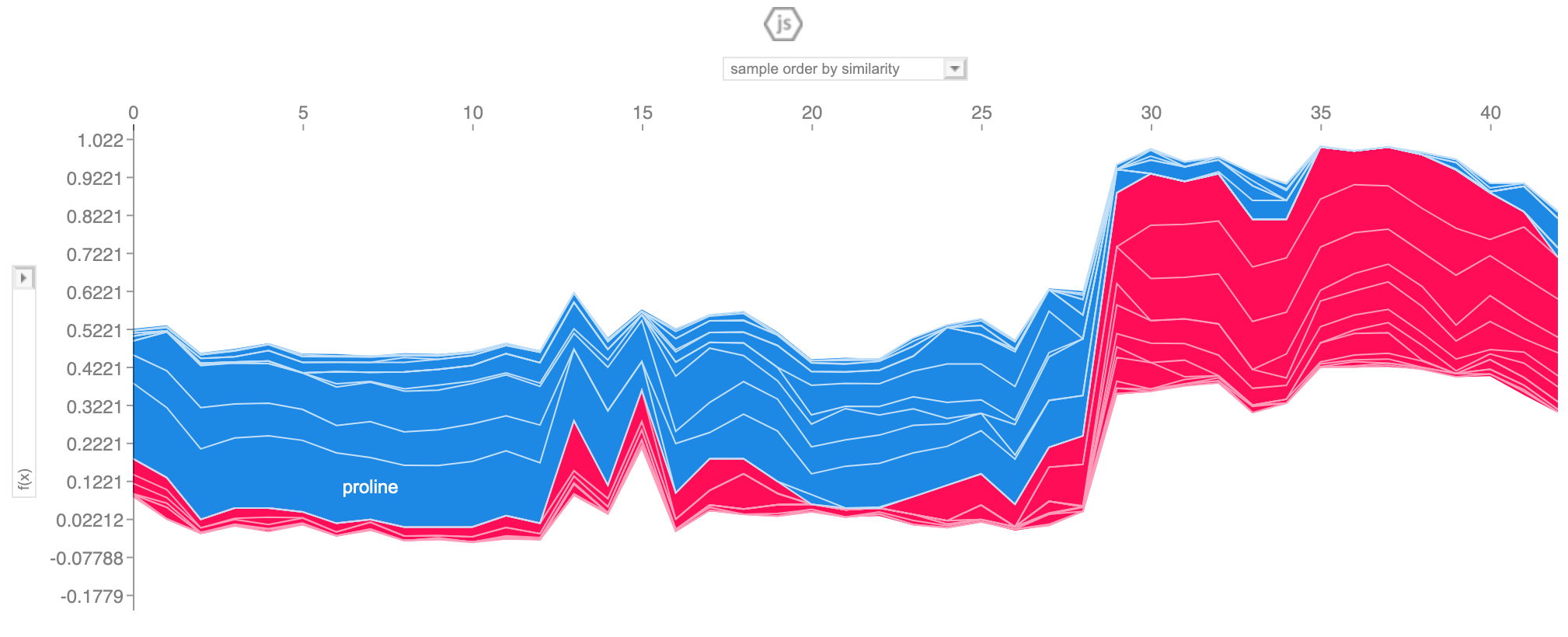
Reason plot
All observation
interpret_model(rf, plot='reason')
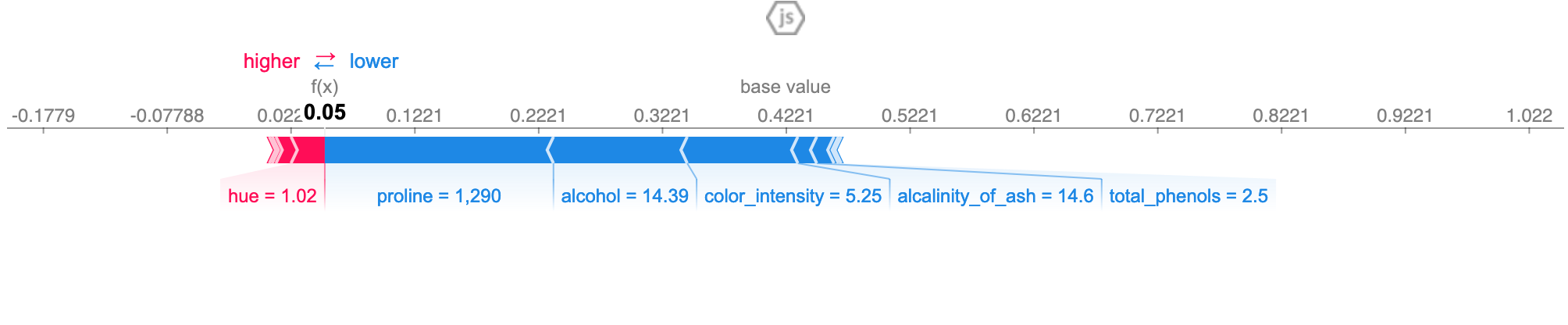
A specific observation
interpret_model(rf, plot='reason', observation=10)
Predicting
Now that the exploratory phase is over, we can predict the results on unseen data.
best_final = finalize_model(best)
predictions = predict_model(best_final, data=df_test.drop('target', axis=1))
from sklearn.metrics import classification_report
print(classification_report(df_test['target'], predictions['Label']))
precision recall f1-score support
0.0 1.00 1.00 1.00 12
1.0 1.00 1.00 1.00 14
2.0 1.00 1.00 1.00 10
accuracy 1.00 36
macro avg 1.00 1.00 1.00 36
weighted avg 1.00 1.00 1.00 36
What now?
PyCaret allows you to do predictions direct from the cloud, so check it out in their website if you want to do this.
Also, you can build your ensemble models with PyCaret.
They support not only Classification, but also Regression, Anomaly Detection, Clustering, Natural Language Processing, and Association Rules Mining.
References
[1] PyCaret Homepage
